Derwent SequenceBase
SequenceBase is a specialized search engine tailored for researchers, patent analysts, and IP professionals working with biomedical patents. The platform enables users to conduct highly targeted searches using biological sequences, delivering precise and relevant patent results.
Problem
At the time, SequenceBase lacked essential features and had unclear, inconsistent functionality, which caused it to fall behind competing platforms in terms of both usability and market competitiveness.
Solution
After identifying key user pain points through research, I implemented critical features all designed to streamline complex workflows and improve search accuracy. These enhancements brought the platform in line with industry standards, making it more competitive and effective for its target audience.
Reshaping the user experience:
Results Table Redesign:
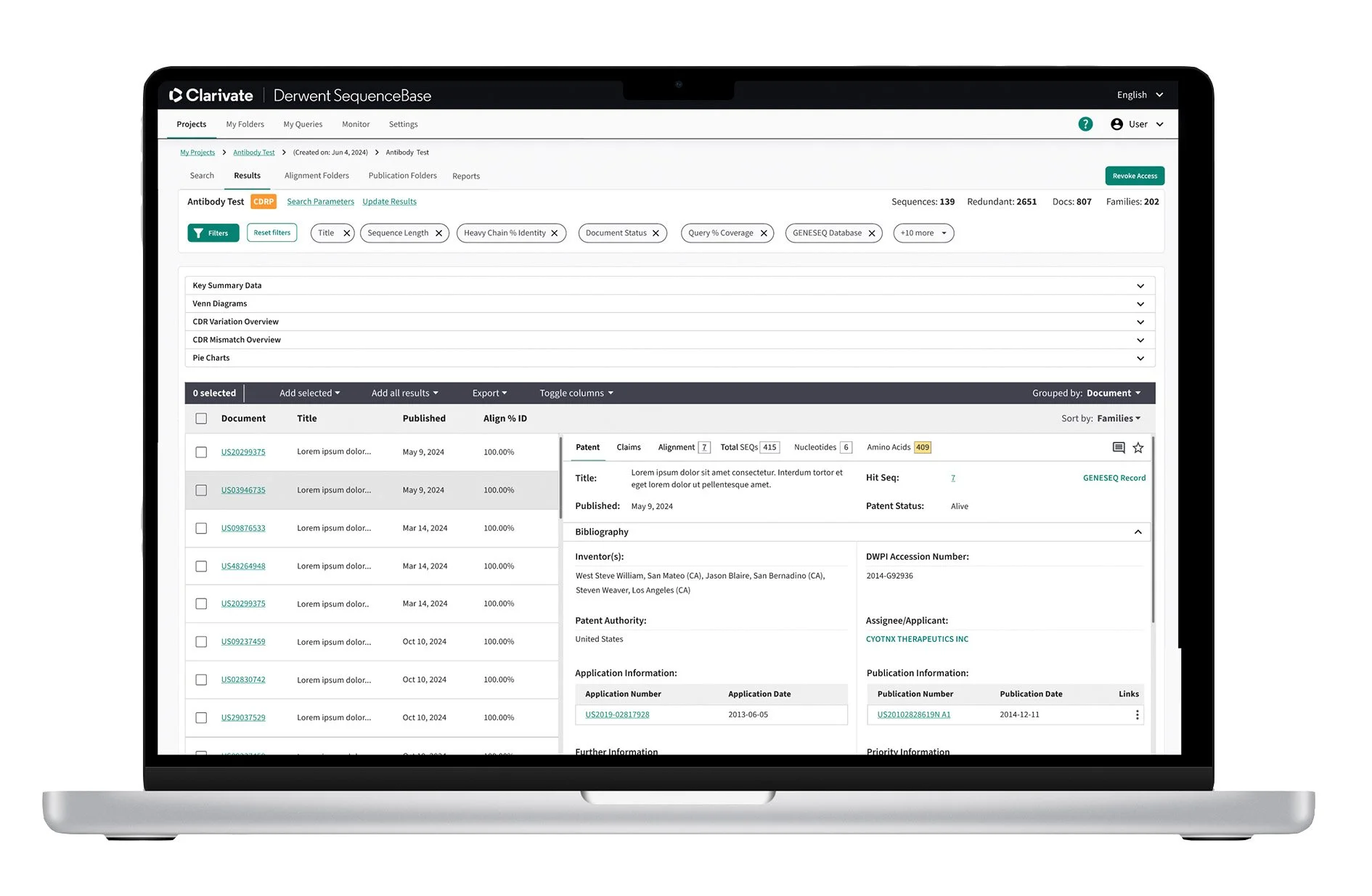
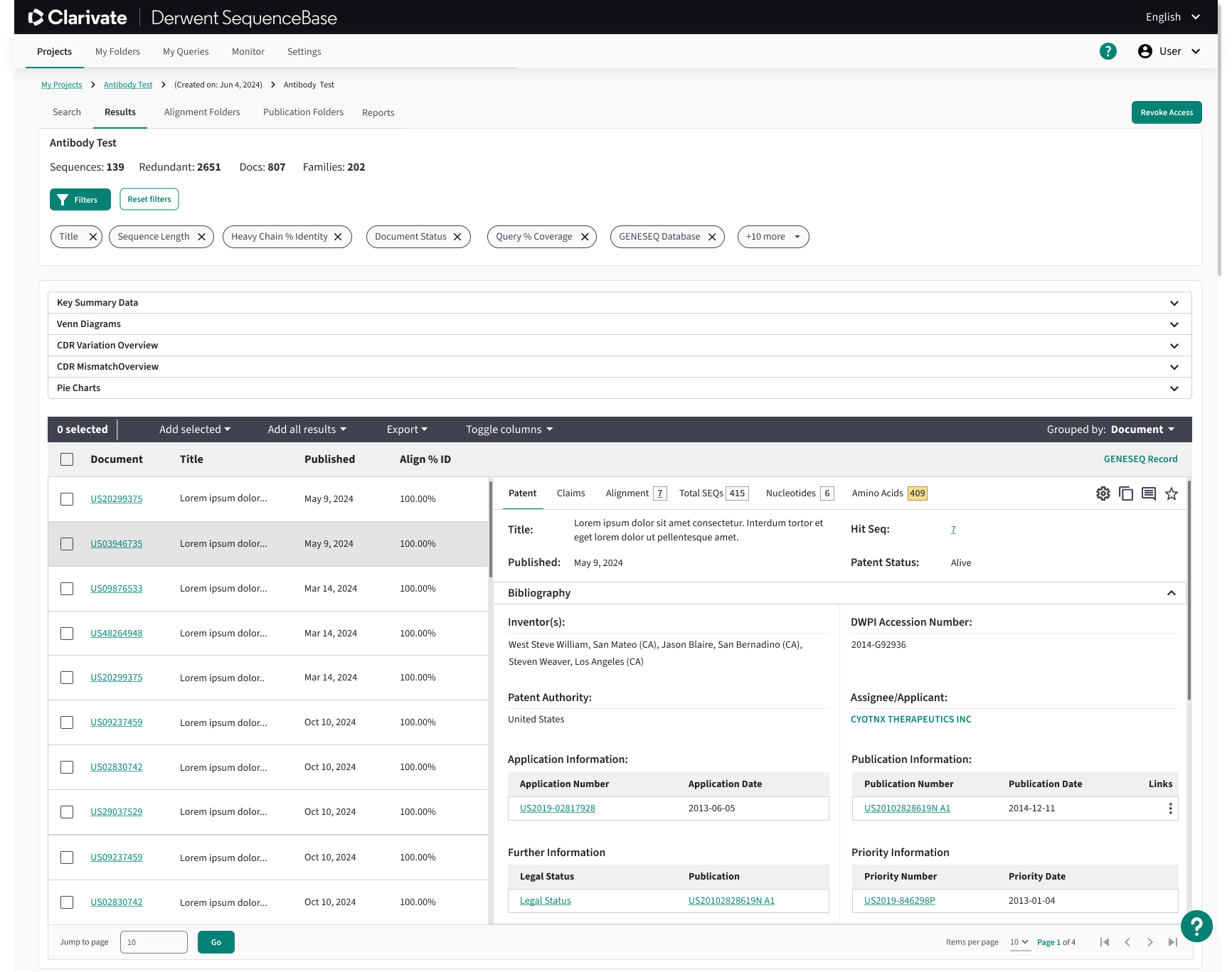
One of the primary challenges with SequenceBase was the results table, which presented significant usability issues. The original design featured overly complex, nested tables with dense, unreadable information that made navigation cumbersome. Once users managed to locate relevant data, they were required to open it in a separate tab, which caused friction in the user flow and forced them to restart the navigation process. This fragmented experience led to inefficiencies and frustration, particularly when users needed to compare or analyze multiple results simultaneously.
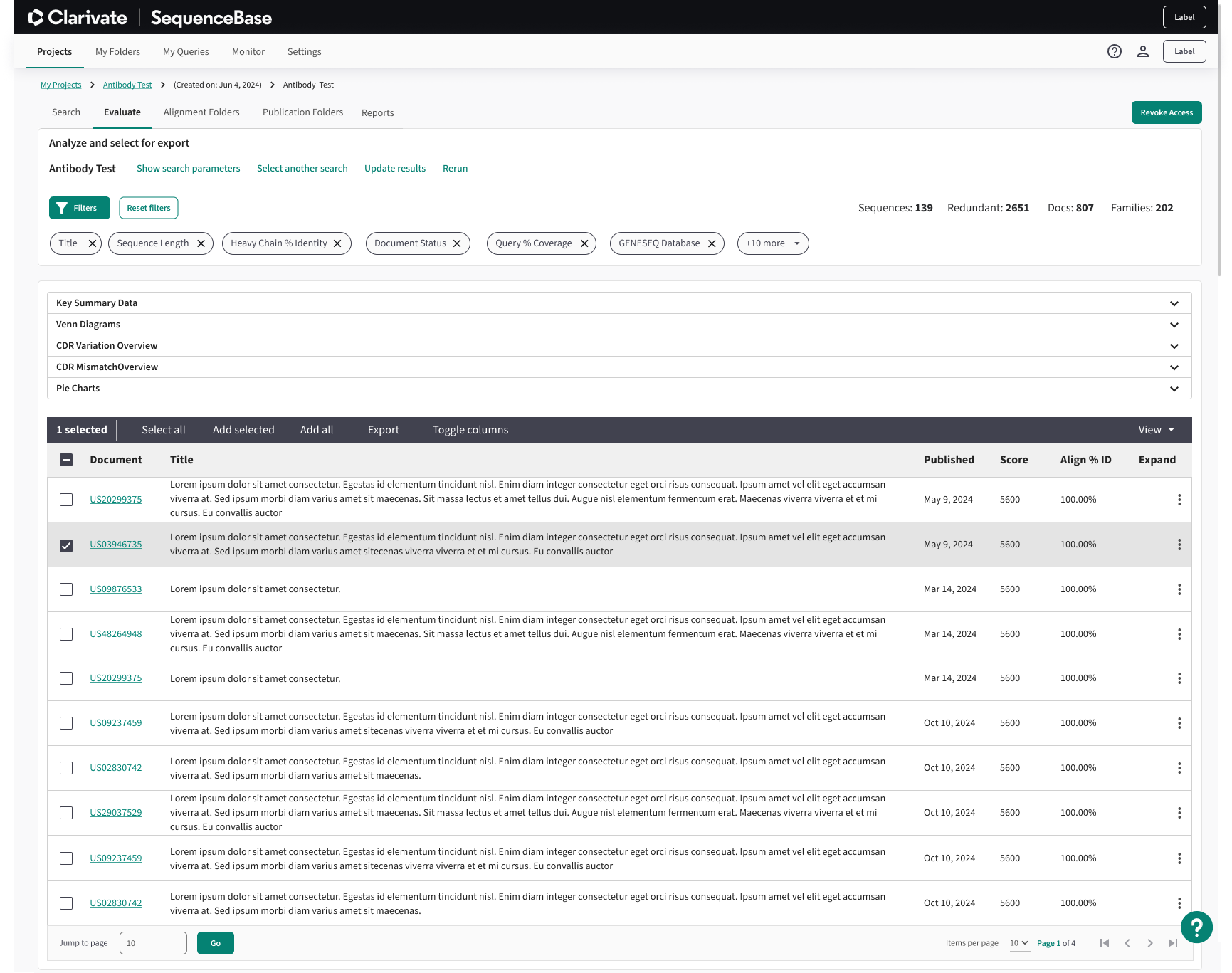
Results Table Solution:
To address the inefficiency caused by managing multiple open tabs, I designed a responsive two-pane results table that consolidates search results and detailed patent information into a single view. The left pane enables users to quickly scan and scroll through relevant patents, while the right pane dynamically displays detailed information for the selected result. This layout reduces context switching, lowers cognitive load, and supports faster comparison and evaluation of patents within a streamlined workflow.
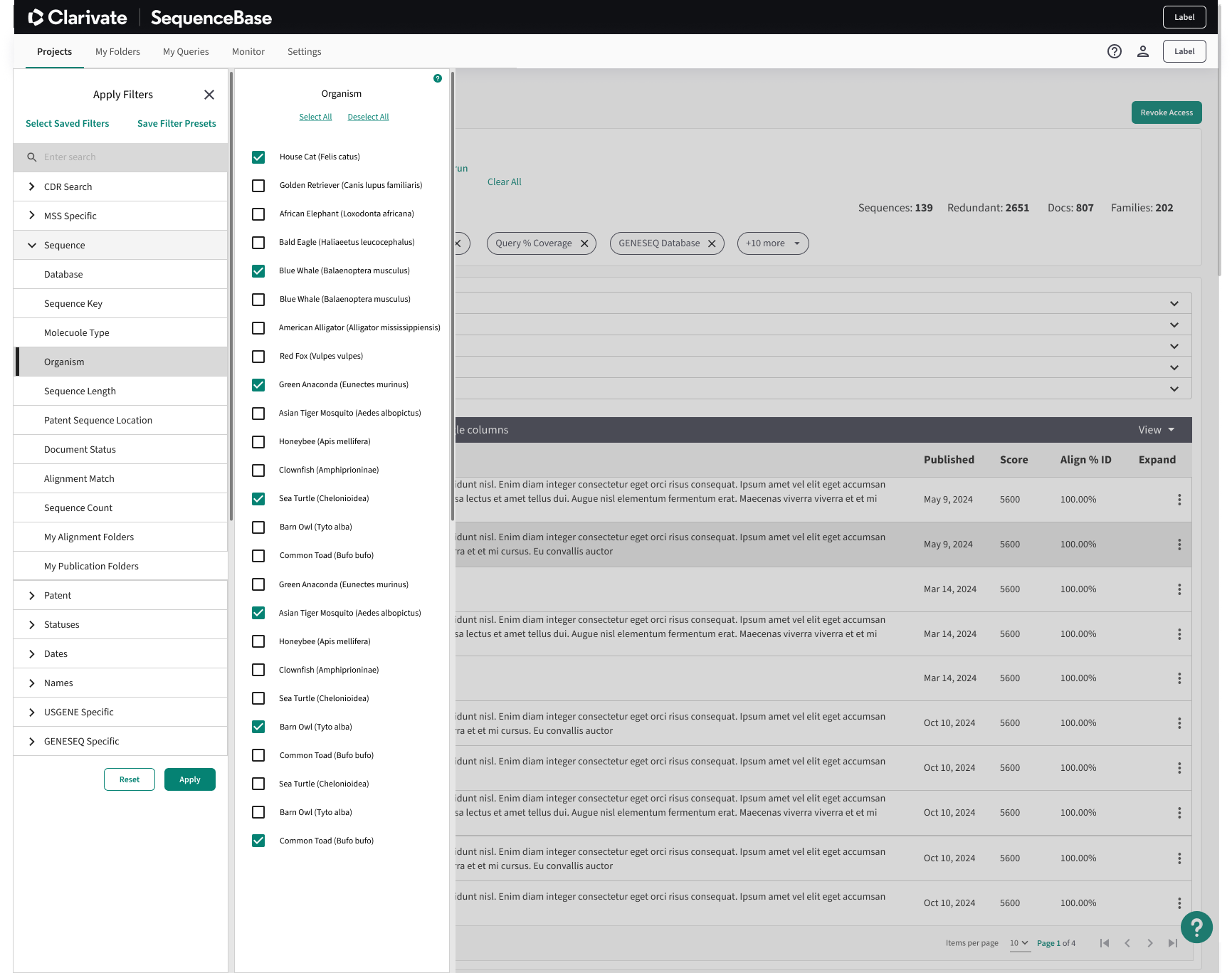
New Filtering Capabilities:
The filtering experience in SequenceBase was a major usability challenge, as it deviated from standard user expectations and mental models. Users were confronted with endless dropdown menus, disorganized lists, and incomplete Venn diagrams, which made it difficult to effectively refine search results. This cluttered and unintuitive interface hindered users from easily accessing the relevant information they needed, impacting the overall efficiency of their workflow.
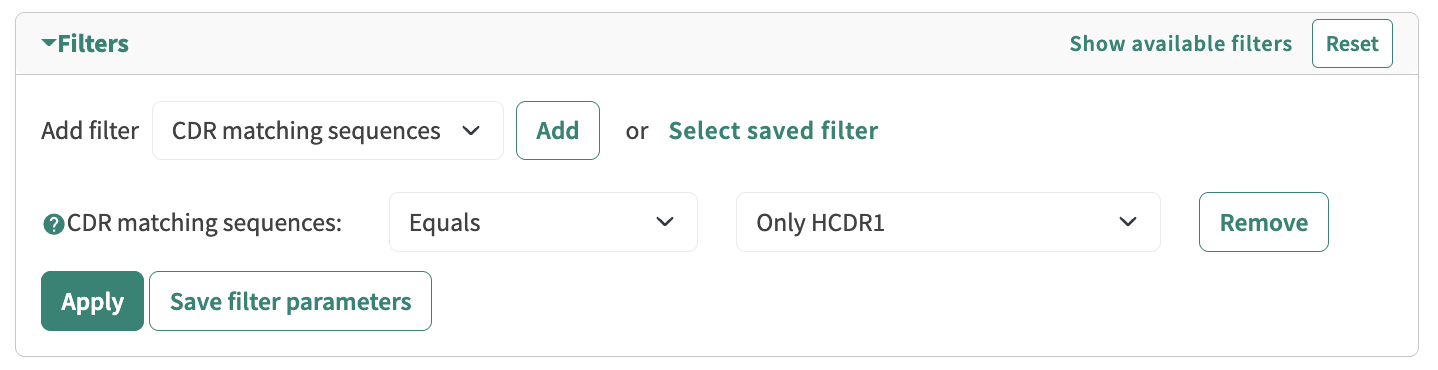
Filtering Solutions:
The first improvement to the filtering system involved transitioning to a more intuitive, menu-style overlay. This design featured neatly organized filter categories, a quick search function for easier navigation, and filter chips for clear, dynamic selection or removal. By streamlining the interface and grouping related options logically, the new design provided users with a more efficient and user-friendly way to refine their searches.
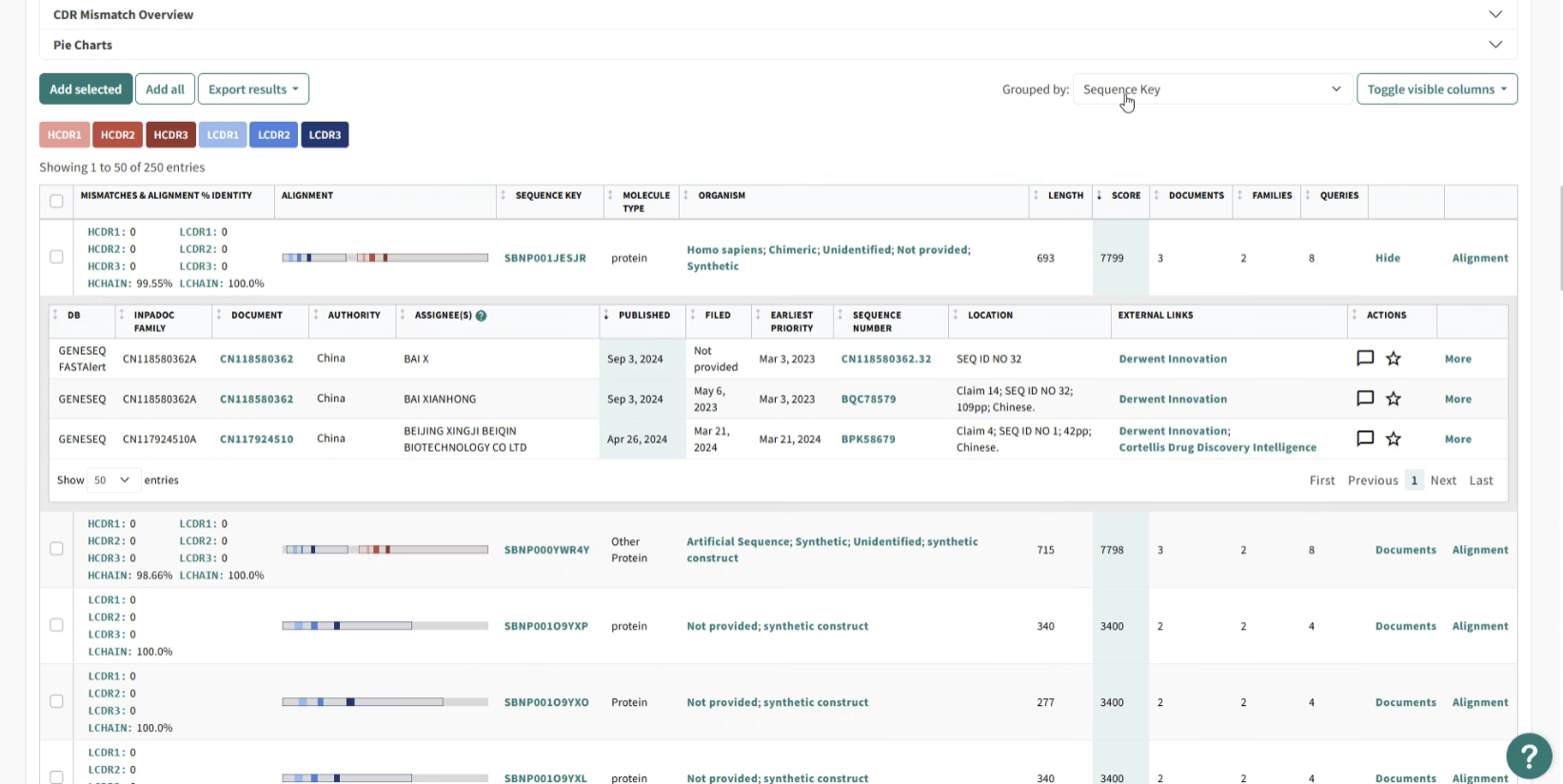
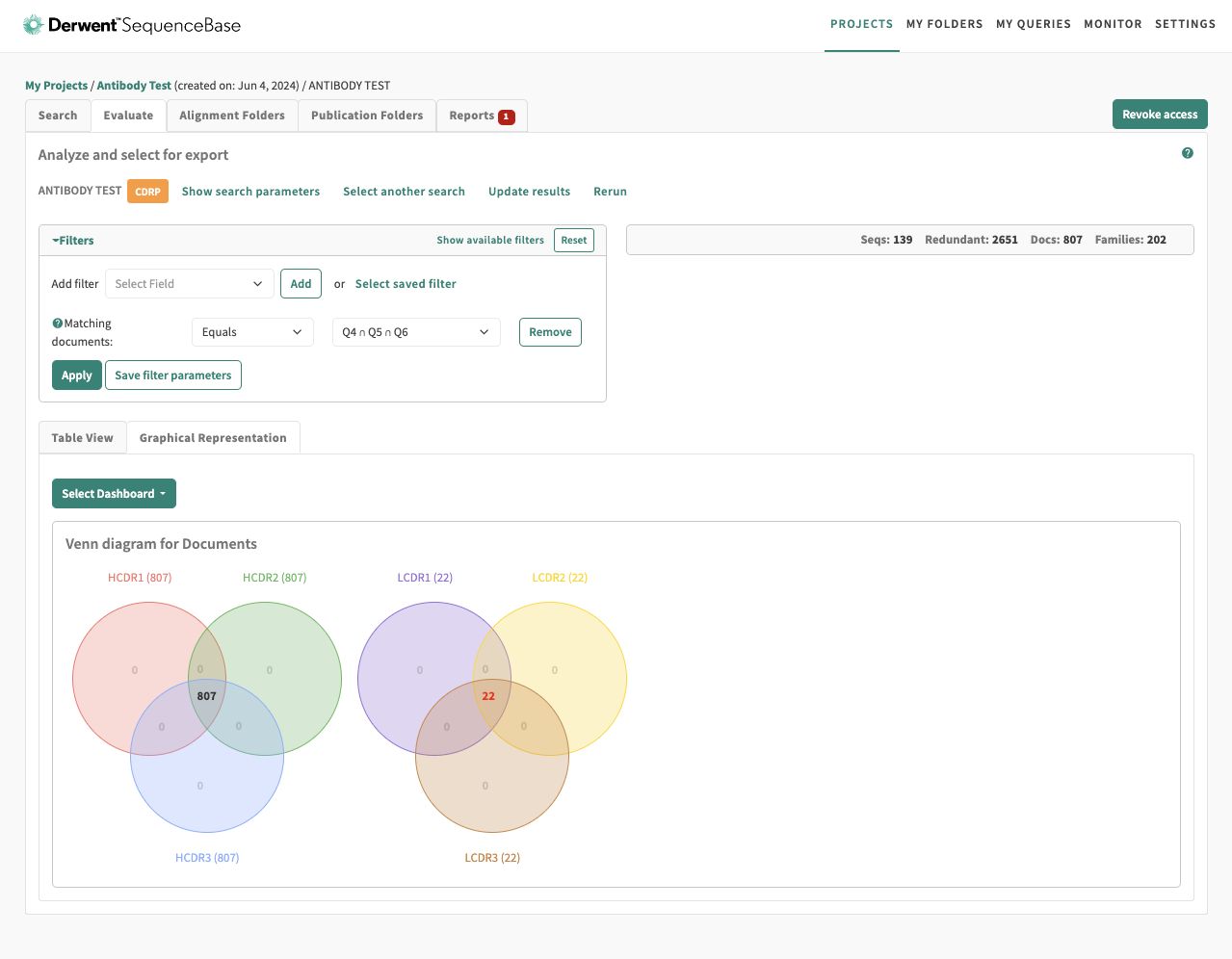
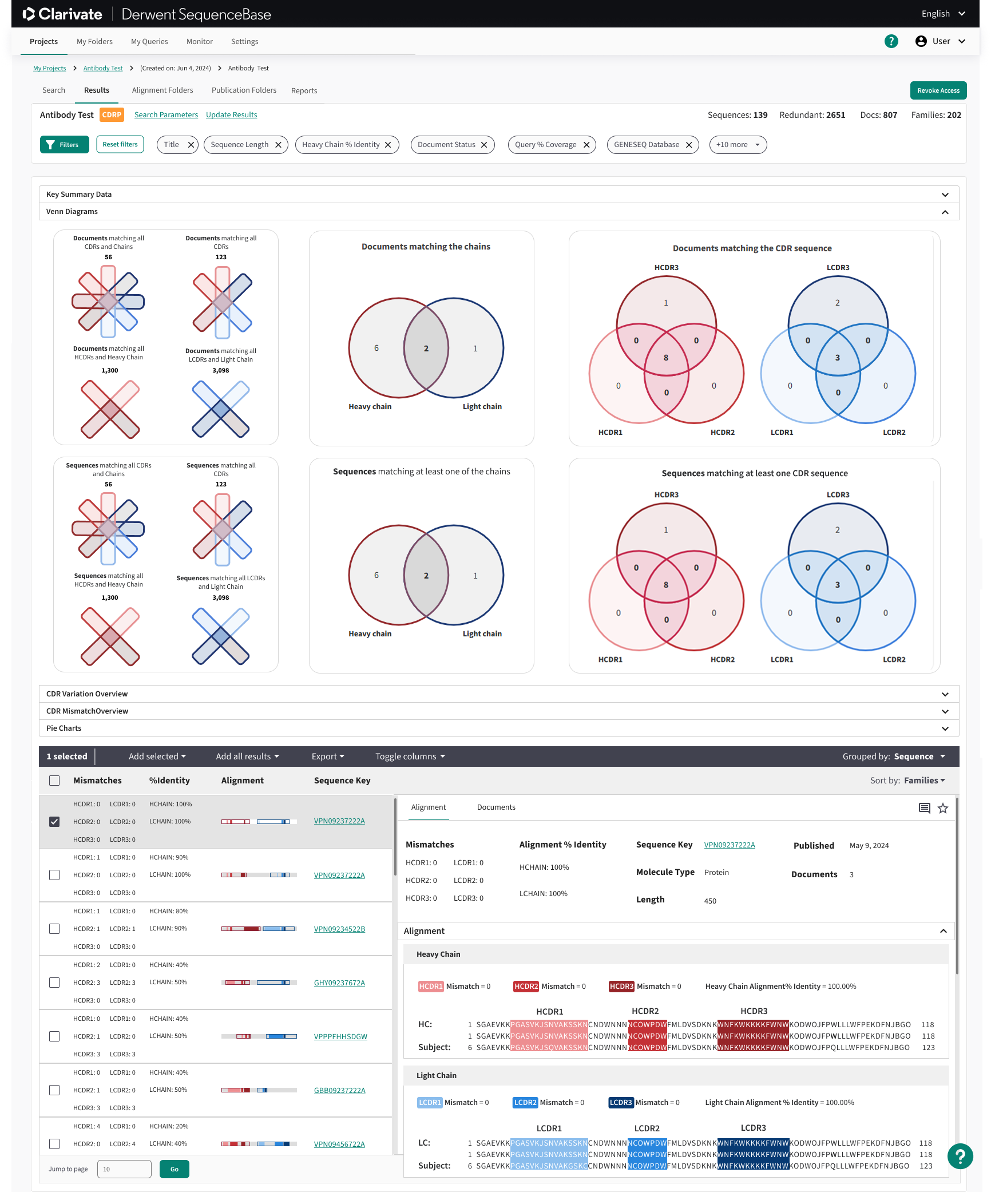
Venn Diagrams And Sequence Highlighting:
In addition to enhancing the standard filtering functionality, we implemented interactive Venn diagrams to further refine search results. These Venn diagrams were fully clickable, allowing users to easily select and combine filter criteria across multiple sections. This addition significantly improved the filtering process, especially for antibody searches, where users needed to visualize complex relationships between multiple datasets.
Additionally, we integrated sequence highlighting into the Venn diagrams, allowing users to visualize the exact sections of genetic sequences they were searching for. This feature was particularly valuable, as genetic sequences can often be extremely long—sometimes spanning tens of thousands of base pairs. By visually emphasizing the relevant sections, we enabled users to quickly identify and focus on the most pertinent data, enhancing the overall efficiency of their searches.
Adding New Functionality:
Sequence Variation Module:
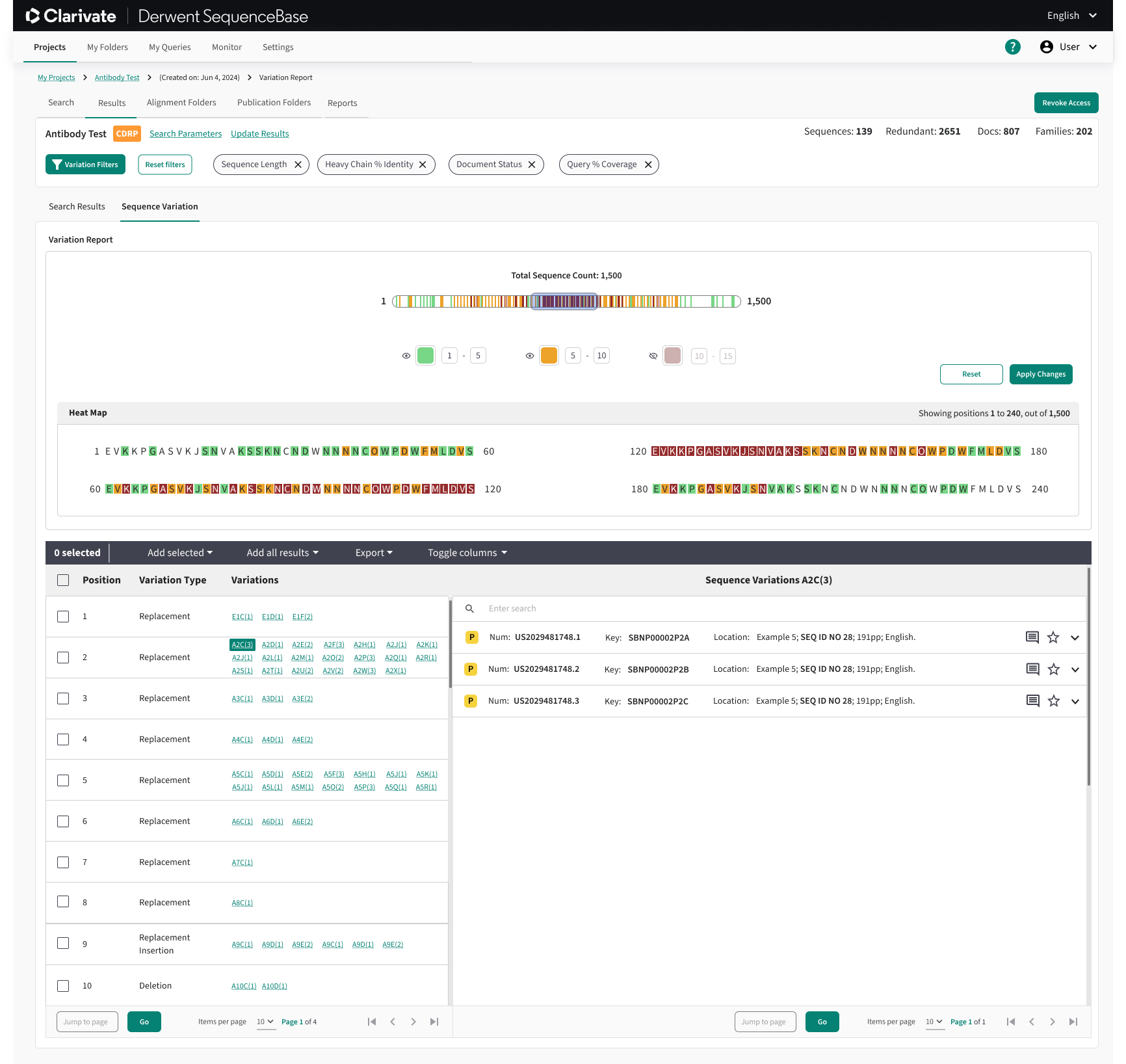
One of the key innovations we introduced to SequenceBase was the Sequence Variation feature, which allowed users to input a query sequence and visualize a heatmap of genetic variations across their search results. This functionality provided a powerful way to identify regions of sequences with high variability, enabling users to include or exclude sequences based on subtle differences in proteins, nucleotides, and amino acids.
To enhance usability, the Sequence Variation module included a dynamic heatmap for clear visualization, a slider window to magnify specific regions for closer examination, and customizable options to hide, show, or adjust the color of variation ranges. These features empowered users to make more informed decisions by allowing for detailed exploration of sequence differences, improving both the precision and flexibility of their searches.
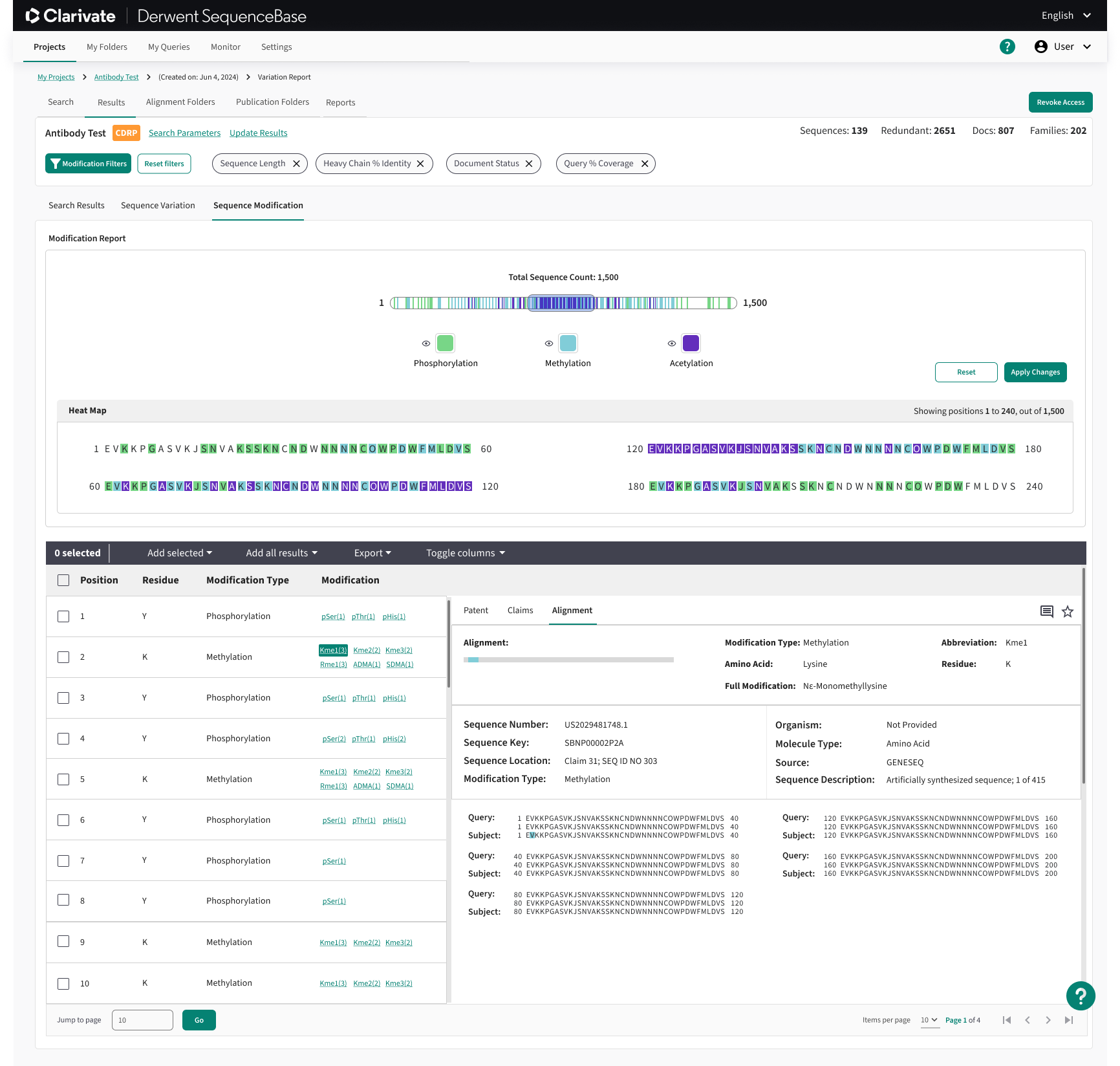
Sequence Modification Module:
The Modification Module operates similarly to the Sequence Variation feature, but instead of visualizing genetic sequence variations, it focuses on the specific chemical modifications or residues that drive those variations. For instance, if protein A in a sequence is altered to protein B due to a methylation or another chemical modification, users can view all patented sequences containing that same chemical change at the same position.
In addition to this, the module includes bonus functionality that allows users to perform searches even without a genetic sequence. By simply entering the sequence length and the chemical residues present in the sequence, users can generate relevant results, offering an efficient way to identify sequences based on chemical modifications alone.
In Conclusion:
By addressing key usability issues in SequenceBase, we significantly enhanced the user experience, making it easier for researchers and patent professionals to search, compare, and analyze genetic sequences. Through the introduction of dynamic results tables, intuitive filtering systems, interactive Venn diagrams, and advanced modules like Sequence Variation and Modification, we not only streamlined complex workflows but also empowered users to make more informed, precise decisions. These improvements reduced friction, increased efficiency, and aligned the interface with users’ mental models, ensuring that SequenceBase is now a more powerful and accessible tool for its target audience.
Ultimately, these design changes not only improved usability but also helped optimize the platform for more focused, accurate, and actionable patent analysis, solidifying SequenceBase as a leading tool in the biomedical patent space.